-Search query
-Search result
Showing all 25 items for (author: jette & ca)

EMDB-13670: 
isolated S-layer of Ca.M.Lanthanididphila
Method: subtomogram averaging / : Gambelli L, Mesman R

EMDB-13672: 
Sub-tomogram average of Ca. M.lanthanidiphila S-layer obtained from whole cell cryo-tomograms
Method: subtomogram averaging / : Gambelli L, Mesman R

EMDB-24504: 
Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C118 (State 1)
Method: single particle / : Barnes CO, Jette CA, Bjorkman PJ

EMDB-24505: 
Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C118 (State 2)
Method: single particle / : Barnes CO, Jette CA, Bjorkman PJ

EMDB-11860: 
Subtomogram average structure of anammoxosomal nitrite oxidoreductase
Method: subtomogram averaging / : Dietrich L

EMDB-11861: 
Helical reconstruction of nitrite oxidoreductase from anammox bacteria
Method: helical / : Parey K

EMDB-23462: 
Structure of CD4 mimetic BNM-III-170 in complex with BG505 SOSIP.664 HIV-1 Env trimer and 17b Fab
Method: single particle / : Jette CA, Bjorkman PJ

EMDB-23465: 
Structure of CD4 mimetic M48U1 in complex with BG505 SOSIP.664 HIV-1 Env trimer and 17b Fab
Method: single particle / : Jette CA, Bjorkman PJ

EMDB-22729: 
Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C002 (state 1)
Method: single particle / : Barnes CO, Malyutin AG, Bjorkman PJ

EMDB-22730: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the human neutralizing antibody Fab fragment, C002 (State 2)
Method: single particle / : Malyutin AG, Barnes CO, Bjorkman PJ

EMDB-22731: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the human neutralizing antibody Fab fragment, C104
Method: single particle / : Barnes CO, Malyutin AG, Bjorkman PJ

EMDB-22732: 
Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C110
Method: single particle / : Dam KA, Barnes CO, Bjorkman PJ

EMDB-22733: 
Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C119
Method: single particle / : Sharaf NG, Barnes CO, Bjorkman PJ

EMDB-22734: 
Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C121 (State 1)
Method: single particle / : Abernathy ME, Barnes CO, Bjorkman PJ

EMDB-22735: 
Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C121 (State 2)
Method: single particle / : Abernathy ME, Barnes CO, Bjorkman PJ

EMDB-22736: 
Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C135
Method: single particle / : Barnes CO, Bjorkman PJ

EMDB-22737: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the human neutralizing antibody Fab fragment, C144
Method: single particle / : Barnes CO, Esswein SR, Bjorkman PJ

EMDB-4921: 
Density map of GluA2cryst with agonist AMPA, Class 1, compact
Method: single particle / : Krintel C, Dorosz J, Larsen AH, Thorsen TS, Venskutonyte R, Mirza O, Gajhede M, Boesen T, Kastrup JS

EMDB-4875: 
Density map of GluA2cryst in the apo state
Method: single particle / : Krintel C, Dorosz J, Larsen AH, Thorsen TS, Venskutonyte R, Mirza O, Gajhede M, Boesen T, Kastrup JS

EMDB-4920: 
Density map of GluA2cryst with negative allosteric modulator GYKI53655 and competitive antagonist ZK200775
Method: single particle / : Krintel C, Dorosz J, Larsen AH, Thorsen TS, Venskutonyte R, Mirza O, Gajhede M, Boesen T, Kastrup JS

EMDB-4922: 
Density map of GluA2cryst with allosteric modulator GYKI53655 and agonist AMPA, Class 2, loose
Method: single particle / : Krintel C, Dorosz J, Larsen AH, Thorsen TS, Venskutonyte R, Mirza O, Gajhede M, Boesen T, Kastrup JS

EMDB-4923: 
Density map of GluA2cryst with negative allosteric modulator GYKI53655
Method: single particle / : Krintel C, Dorosz J, Larsen AH, Thorsen TS, Venskutonyte R, Mirza O, Gajhede M, Boesen T, Kastrup JS

EMDB-4924: 
Density map of GluA2cryst with agonist AMPA, Class 2, loose
Method: single particle / : Krintel C, Dorosz J, Larsen AH, Thorsen TS, Venskutonyte R, Mirza O, Gajhede M, Boesen T, Kastrup JS
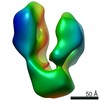
EMDB-4925: 
Density map of GluA2cryst with allosteric modulator GYKI53655 and agonist AMPA, Class 1, compact
Method: single particle / : Krintel C, Dorosz J, Larsen AH, Thorsen TS, Venskutonyte R, Mirza O, Gajhede M, Boesen T, Kastrup JS
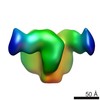
EMDB-8507: 
92BR SOSIP.664 trimer in complex with DH270.1 Fab
Method: single particle / : Fera D, Harrison SC
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
